RESEARCH ARTICLE
caGrid-Enabled caBIGTM Silver Level Compatible Head and Neck Cancer Tissue Database System
Haibin Wang *, 1, Erik Bouzyk 1, Anna Kuehn 2, 3, Susan Muller 2, 3, Zhengjia Chen 1, 6, Fadlo R Khuri 4, Dong M Shin4, André Rogatko 5, Mourad Tighiouart 1, 6
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 171
Last Page: 178
Publisher Id: TOMINFOJ-4-171
DOI: 10.2174/1874431101004010171
Article History:
Received Date: 2/11/2009Revision Received Date: 23/4/2010
Acceptance Date: 21/6/2010
Electronic publication date: 1/9/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
Abstract
There are huge amounts of biomedical data generated by research labs in each cancer institution. The data are stored in various formats and accessed through numerous interfaces. It is very difficult to exchange and integrate the data among different cancer institutions, even among different research labs within the same institution, in order to discover useful biomedical knowledge for the healthcare community. In this paper, we present the design and implementation of a caGrid-enabled caBIGTM silver level compatible head and neck cancer tissue database system. The system is implemented using a set of open source software and tools developed by the NCI, such as the caCORE SDK and caGrid. The head and neck cancer tissue database system has four interfaces: Web-based, Java API, XML utility, and Web service. The system has been shown to provide robust and programmatically accessible biomedical information services that syntactically and semantically interoperate with other resources.
INTRODUCTION
Biomedical research labs have used various tools to generate data of unprecedented depth, timeliness and diversity for scientists and clinicians to attack the problems of preventing and curing human cancer. The diversity of data utilizes heterogeneous storage formats (plain text, Excel spreadsheet, Access database, various SQL databases, etc), different database schemas, metadata, and various interfaces for accessing the data. This diversity impedes the integration and interoperation of disparate data systems created in each research lab for the purposes of performing data analyses of multiple types [1]. The solution to this problem is to build data systems utilizing an architecture that facilitates such interoperability.
In 2003, the National Cancer Institute (NCI) launched the cancer Biomedical Informatics Grid (caBIGTM), an international collaboration to facilitate and enable research teams to share data, applications, and infrastructure that facilitate collaborations that accelerate the conversion of data from information to knowledge [2]. To accomplish this goal, the caBIGTM community is developing standards, policies, guidelines, common applications, open-source tools, and middleware infrastructure to enable more effective sharing of data and research tools among scientists and organizations in a multi-institutional environment [3]. In the latest version (v3.0) of the caBIGTM Compatibility Guidelines [4], four maturity levels of interoperability between systems are described: legacy, bronze, silver, and gold.
Under the leadership of the NCI, its Center for Bioinformatics (NCICB) and caBIGTM participants, many infrastructures, tools, and services (such as: caGrid 1.2 [5], cancer Common Ontological Representation Environment Software Development Kit (caCORE SDK) [6], Semantic Integration Workbench (SIW), and UML Model Loader 4.0 [7]) have been implemented to help design, develop, and deploy data and analytical services with full syntactic and semantic interoperability.
caGrid is an underlying Grid middleware infrastructure for caBIGTM to support fully syntactic and semantic interoperability among resources through a federation of such resources [8]. caGrid is a model-driven and service-oriented architecture that synthesizes and extends a number of technologies such as Globus Toolkit, Mobius Global Model Exchange (Mobius GME), cancer Data Standards Repository (caDSR) [9], Enterprise Vocabulary Services (EVS) [10], ActiveBPEL, Grouper, etc. to provide a standardized framework for the advertising, discovery, and invocation of data and analytical resources [3]. The latest release version is caGrid 1.2 [5]. caGrid 1.2 provides the common grid infrastructure upon which the caBIGTM silver level compliant grid services and tools are built.
caCORE is an interoperability infrastructure based on Model Driven Architecture (MDA), n-tier architecture, and a common application programming interface (API) for data access [1]. Biomedical data systems built using the caCORE fulfill both aspects of interoperability: exchanging data (syntactic interoperability) and processing the exchanged data (semantic interoperability) [11]. The caCORE infrastructure consists of an integrated set of three components: a controlled terminology service (the EVS), a standards-based metadata repository (the caDSR), and an information system with an API based on domain MDA [1].
The term “head and neck cancer” refers to a group of biologically similar cancers originating from the upper aerodigestive tract, including the lip, oral cavity (mouth), nasal cavity, paranasal sinuses, pharynx, and larynx. Head and neck cancer is highly curable if detected early. The effective treatment for head and neck cancer is usually some kind of surgery, but chemotherapy and radiation therapy may also play important roles [12].
We designed and implemented the head and neck cancer tissue database (HNCTD) system following the cancer Biomedical Informatics Grid (caBIGTM) paradigm. The system fully supports syntactic and semantic interoperability. It is caBIGTM silver level compatible according to caBIGTM compatibility guidelines [4]. The HNCTD is an Internal Review Board (IRB)-approved Health Insurance Portability and Accountability Act (HIPAA) compliant data system. Currently, the HNCTD contains information on 209 cases of head and neck squamous cell carcinomas (HNSCC) from 1996-2003. All tissues in the database are formalin fixed, paraffin embedded tissues housed at either Emory University Hospital or Emory Midtown Hospital. Information from consented patients is extracted from the Surgical Pathology files in the Department of Pathology and Laboratory Medicine [13].
The information in the HNCTD includes date of birth, date of diagnosis, tumor site, tumor stage and grade, smoking history, radiation treatment, chemotherapy treatment, date of recurrence, and date of last follow up or date of death. In addition, if the patient developed a second primary tumor the related information was also documented. The database is divided into two groups: no metastases (NoMET) and metastases (MET).
METHODS
We designed and implemented the head and neck cancer tissue database system based on the caBIGTM paradigm. We utilize the open source software such as ArgoUML (the UML modeling tool) [14], caCORE SDK 4.0, caGrid 1.2, and Introduce (the caGrid service creation toolkit) [15], etc. The process flow for the system design is shown in Fig. (1).
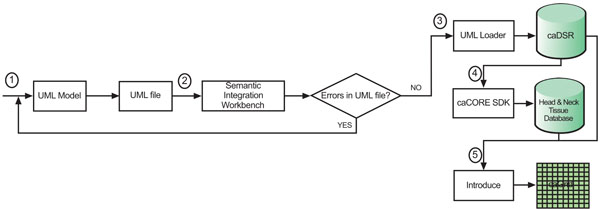 |
Fig. (1). Process flow for the system design. |
The algorithmic steps for the process flow are described as:
Begin:
Step 1: Build the object and data models using a Unified Modelling Language (UML) modeling tool such as Enterprise Architect [16] or ArgoUML
Step 2: There are several separate steps in running Semantic Integration Workbench (SIW):
Step 2.1 (by Model Owner): Review unannotated XML Metadata Interchange (XMI) or UML file built in Step 1.
Step 2.2 (by Model Owner): Perform XMI or UML roundtrip.
Step 2.3 (by Model Owner): Run semantic connector.
If no errors such as invalid data types and “unbounded array” in model found in steps 2.1-2.3, then proceed to step 2.4; otherwise, go to step 1 to correct the errors and repeat steps 2.1-2.3.
Step 2.4 (by Vocabulary Reviewer): Send XMI or UML file via email to the NCICB to curate the file.
Step 2.5 (by Model Owner): Review annotated XMI or UML file.
Step 2.6 (by Model Owner): Generate default Global Model Exchange (GME) tags.
Step 2.7 (by Model Owner): GME cleanup.
If no errors found in steps 2.4-2.7, then proceed to step 3; otherwise, go to step 1 to correct the errors and repeat steps 2.1-2.7.
Step 3: Run UML Loader by the NCICB to load the approved annotated XMI or UML file into the caDSR which can be accessed at runtime.
Step 4: Run the caCORE SDK with the approved annotated XMI or UML file to generate the ‘caCORE-like’ data system to provide ‘strongly typed objects’ and object relational mapping access to the backend head and neck cancer tissue database.
Step 5: Run Introduce [16] with the approved, annotated XMI or UML file and generated caCORE-like system artifacts to define the grid data service interface over the object oriented APIs that define functionality exposed to the caGrid.
End.
According to step 1, we first need to create the object and data models for the underlying database system. Here, we use ArgoUML 0.26, a freeware application, to create the models. The output of this procedure is a UML (XML format) file. As we mentioned before, the head and neck cancer tissue database is divided into two groups: metastasis and non-metastasis. Therefore, we model domain objects in two classes. Screenshots of the object and data models in ArgoUML 0.26 are shown in Figs. (2, 3).
 |
Fig. (2). Object models of the HNCTD. |
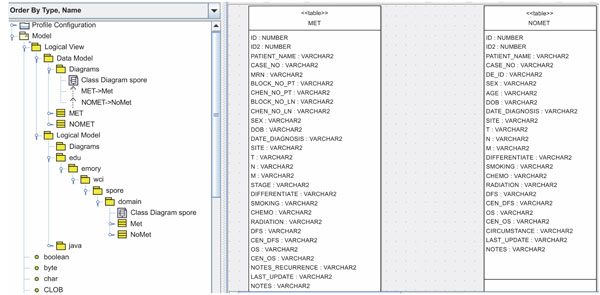 |
Fig. (3). Data models of the HNCTD. |
One design requirement of the HNCTD is that principle investigators (PIs) and researchers who use the system can add new columns (biomarks) to the existing MET and NoMET tables, which means the database schema changes over time. In next subsection, we describe the principle of a semi-automatic time variant schema-enabled UML file generator. Basically, it is a reengineering process to get new domain object and data models using existing database schema.
Semi-Automatic Time Variant Schema-Enabled UML File Generator
We are able to automate the generation of the UML file each time the HNCTD schema is modified. A script is run on the web server at intervals specified by the administrator. The script queries the database and returns every column. The query is then compared to a previous query to check for any changes in the database’s state. If a change has been detected, the script modifies the UML file appropriately and notifies the administrator of the change via e-mail. The administrator can run the SIW to check the new UML file and submit it to the NCICB for annotation and loading into the caDSR. The UML file in use by the caCORE SDK is replaced by the modified UML file so that access to up-to-date data is always available. The process flow of the semi-automatic time variant schema-enabled UML file generator is shown in Fig. (4). The script is available for downloading at: http://sisyphus.emory.edu/UML_generator.zip.
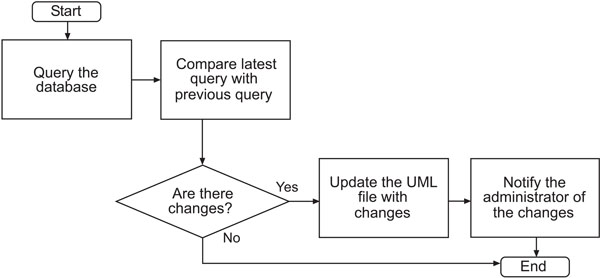 |
Fig. (4). Flowchart of UML file generator. |
System Architecture
Because the database contains identifiable patient information, such as patient’s name, medical record number, date of birth, etc., we prioritize the security of the whole data system to make it a HIPAA compliant. We integrate the Common Security Module (CSM) [16], Grid Authentication, and Authorization with Reliably Distributed Services (GAARDS) [17] into the system architecture. The high level architecture of the caGrid-enabled caBIGTM silver level compatible HNCTD system is shown in Fig. (5).
As we mentioned before, we apply the caCORE SDK 4.0 with CSM enabled to our approved annotated model to generate a ‘caCORE-like’ data system. The generated data system only supports querying (to be discussed in Results section), but not data input (to be discussed in Results section). Therefore the data system is read-only.
In Fig. (5), there are three workflows (red line, green line, and orange line) to support the web-based user interface and one workflow (blue line) to support the caGrid data service API.
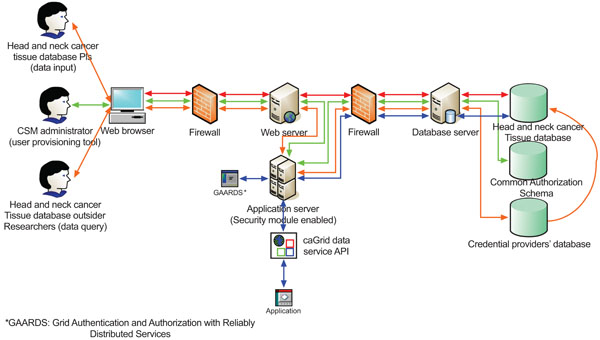 |
Fig. (5). High level architecture of the HNCTD system. |
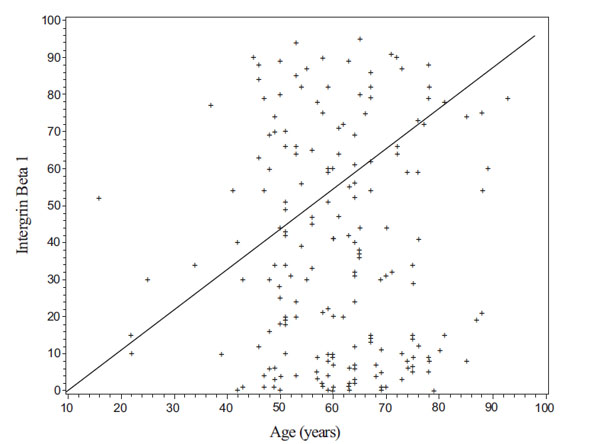 |
Fig. (6). A scatter plot of patient’s Intergrin Beta 1 value by his/her age in years. |
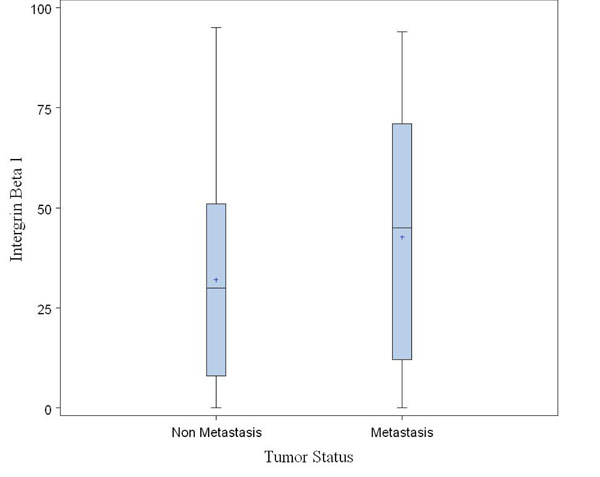 |
Fig. (7). A boxplot for comparison of Intergrin Beta 1 between non metastasis and metastasis patients. |
- Workflow 1 (red line): The PIs and researchers of the head and neck cancer Specialized Programs of Research Excellence (SPORE) [18] are internal users of the database system. They can interact with the web browsers to retrieve the archived head and neck cancer tissue data and input new tissue information. Results section will describe the design of this data input interface.
- Workflow 2 (green line): The CSM administrator utilizes the User Provisioning Tool (UPT) [19], a web-based management tool for authentication and authorization of users of the underlying database system, to set up credentials and different levels of access privileges for data, methods, and objects for external users. The CSM uses the credential provider’s database to authenticate and the common authorization schema and database to authorize.
- Workflow 3 (orange line): The head HNCTD system’s external users use web browsers to query the tissue data. Firstly, the system will interact with the integrated CSM for authentication. If a user’s credentials are valid, the user can query the database according on assigned access privileges.
- Workflow 4 (blue line): The HNCTD system also provides the caGrid data service API for third party applications to access the database with GAARDS-enabled security control. This data service interface supports fully syntactical and semantic integration among disparate systems.
RESULTS
Data Input Interface
In order for the designed database system to be useful, we provide a data input interface. Because the caCORE SDK 4.0 does not support an API for writing data, we choose to design and implement the data input subsystem separately.
The data input subsystem is designed with collaboration and security in mind. There are two kinds of users of this subsystem: system administrators and common users (clinicians, researchers, and biostatisticians). Administrators are able to customize each user’s specific levels of access (such as the addition of new columns, and the reading, writing, and deletion of certain fields and patient data). This enables researchers to view necessary data and allows for the ability to share information while maintaining patient’s privacy. Users are able to export their data (in a tab-delimited text file and/or an excel file) for offline use. Users are also able to do the simple search by patient’s name, medical record number (MRN), Case Number, or De-Identified Number. The data input interface is accessible at https://wci9.cc.emory.edu/spore.
The technical specifications for our data input subsystem are provided in Appendix 1.
Here we provide a successful application example of research project using our database. The project is to study the difference in a biomarker, Intergrin Beta 1, between non metastasis and metastasis patients. A dataset consisting of 101 metastasis and 106 non metastasis patients is collected using our database and exported by a user (Z. Chen). Some of analysis results with the dataset, such as a scatter plot of patient’s Intergrin Beat 1 by his/her age and a boxplot for comparison of Intergrin Beta 1 between non metastasis and metastasis patients, are shown in Figs. (6, 7), respectively.
Data Query Interface
We developed the HNCTD data query subsystem using caCORE SDK 4.0 with CSM enabled. The generated system has four kinds of user query interfaces [20]: (1) Web-based, which helps users view data in the form of a web page with a web browser. The CSM-enabled web-based query interface is accessible at: http://velda.emory.edu:8081/spore.; (2) XML utility-based, which facilitates thin clients to communicate with the generated data system with an XML-based REST interface; (3) Java API-based, which enables a Java program to be developed with this API to communicate with the generated data system; (4) Web service-based, which allows an application to be built by calling the Web service API to communicate with the generated ‘caCORE-like’ data system.
Besides these four interfaces, we also use Introduce 1.2, an integrated tool from caGrid 1.2 to create and deploy a GAARDS-enabled caGrid data service for the caCORE SDK generated data system. For detail steps for creating, deploying, and invoking the data service please refer to the Introduce tutorial [21].
The technical specifications for data query subsystem are provided in Appendix 2.
DISCUSSION
To further improve treatment for cancer patients, it is necessary to conduct translational studies which link clinical outcomes with tissue based molecular data and unravel the mechanism of cancer in the molecular point of view. Some of the challenges in contemporary cancer translational research include, but not limited to, data sharing, data complexity, and organizational complexity. A sharing database built with the caBIGTM paradigm is the answer for these challenges. Our database is established with the core of caBIGTM paradigm, such as the XML, model driven architecture, object-relational mapping, semantic web, and grid computing technologies. Therefore, our database is easy to upgrade in the future and a similar database as ours can be easily established in other institutes according to their needs. In the market, caTissue is a good alternative caBIGTM’s biorepository tool for biospecimen inventory management, tracking, and annotation. This tool enables users to contribute and retrieve data for the purpose of tissue storage, quality assurance, and distribution of biospecimens [22]. caTissue suite is open source, but its full service support requires high license fee.
Our HNCTD system mainly stores head & neck cancer clinical information. The system is designed and developed according to caBIGTM paradigm. It facilitates the data sharing based on the open standards and has a user-friendly graphical user interface (GUI) to help access the patient data in the database. Because the HNCTD stores patient’s identity and private information, we put a lot of efforts for the security of the system in order to satisfy the HIPAA requirements. The HNCTD system also has some limitations. For example, it does not support the storage and retrieval of biospecimen such as image; it cannot integrate with the OnCore clinical system running in Winship Cancer Institute of Emory University; and it has not been integrated with caTissue Suite running in Emory University. But we are endeavoring to overcome these limitations by redesigning the system with caCORE SDK 4.1 after a future release of caGrid is available.
CONCLUSION
The HNCTD system is developed based on the caBIGTM paradigm. It demonstrates the effectiveness of rapid application development for building ‘caCORE-like’ data system using model driven architecture (MDA) and n-tier distributed computing architecture methodology. The system is built with a set of open source applications that are easily accessible to any application development team. We deploy our caCORE SDK-generated data system on the caGrid infrastructure using the Introduce toolkit to make the whole system caBIGTM silver level compatible. The HNCTD system realizes the syntactical and semantic interoperability among multisite which host the head & neck tissue data. In addition, the system has a lot of special features, such as: reporting, user management, logging, and adding/deleting new biomarkers.
Currently, the HNCTD is just open to the SPORE PIs, researchers, and biostatisticians. For the external users, he/she should register first and wait for the system administrator’s approval. In the future, we are going to advance the HNCTD by developing analytical services for the data mining of stored head and neck cancer tissue information. Also, we will develop a sophisticated time variant schema-enabled UML file generator for more general user case. We will also redesign the system to adopt caCORE SDK 4.1 (for the support of an API which enables data input) once we obtain a future release of caGrid.
ACKNOWLEDGEMENTS
The authors thank Karen Lyn Exline and Belinda Maaskant for their assistance in configuring the web and application servers, as well as Kenneth Buck for his assistance in setting up the Oracle 10g database. This project is supported by the NIH/NCI – 5 P50 CA128613 (SPORE in Head and Neck Cancer).
DISCLOSURE
The authors report no conflicts of interest.
APPENDIX
APPENDIX 1
The technical specifications for our data input subsystem:
1. Web Server
- Red Hat Enterprise Linux
- Apache 2
- PHP 5 engine
- SSL-enabled secure HTTP connection
2. Database Server
- Red Hat Enterprise Linux
- Oracle 10g Release 2
3. Programming Language and Library
- PHP 5
- PDO OCI
- GD Library for PHP
- Zend optimizer
APPENDIX 2
1. Web Server
- Red Hat Enterprise Linux
- Apache 2
- SSL-enabled secure HTTP connection
2. Application Server
- Tomcat 5.x
- JBoss 4.x
3. Database Server
- Red Hat Enterprise Linux
- Oracle 10g Release 2
4. Programming Language and SDK
- J2EE
- Java SDK 1.5.x
- Apache Ant 1.6.5
- ArgoUML 0.26
- caCORE SDK 4.0
- caGrid 1.2
- Introduce 1.2
REFERENCES
| [1] | Komatsoulis G, Warzel D, Hartel F, et al. caCORE version 3: Implementation of a model driven, service-oriented architecture for semantic interoperability J Biomed Inform 2008; 41: 106-23. |
| [2] | Eschenbach AC, Buetow K. Cancer Informatics Vision: caBIGTM Cancer Inform 2006; 2: 22-4. |
| [3] | Saltz J, Oster S, Hastings S, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid Bioinformatics 2006; 22(15): 1910-6. |
| [4] | caBIGTM Compatibility Guidelines Available from: https://gforge. nci.nih.gov/frs/download.php/3948/caBIG_Compatibility_Guidelines_v3.0_Final.pdf |
| [5] | caGrid 1.2 Available from: http://cagrid.org/wiki/CaGrid:Softw are:Release:1.2 |
| [6] | Cancer Common Ontologic Representation Environment Software Development Kit 4.0 (caCORE SDK 4.0) Available from: http://ncicb.nci.nih.gov/download/downloadcacoresdk.jsp |
| [7] | Semantic Integration Workbench (SIW) and UML Loader 4.0 Available from: https://gforge.nci.nih.gov/docman/index.php?. group_id=16&selected_doc_group_id=4199&language_id=1 |
| [8] | Saltz J, Kurc T, Hastings S, et al. e-Science, caGrid, and translational biomedical research IEEE Comput Soc 2008; 41(11): 58-66. |
| [9] | Cancer Data Standards Repository (caDSR) Available from: http://ncicb.nci.nih.gov/NCICB/infrastructure/cacore_overview/cadsr |
| [10] | Enterprise Vocabulary Services (EVS) Available from: http://ncicb.nci.nih.gov/NCICB/infrastructure/cacore_overview/vocabulary |
| [11] | Tobias J, Chilukuri R, Komatsoulis G, et al. The CAP cancer protocols – a case study of caCORE based data standards implementation to integrate with the Cancer Biomedical Informatics Grid BMC Med Inform Decision Making 2006; 6: 25. |
| [12] | Head and Neck Cancer Available from: http://en.wikipedia.org/ wiki/Head_and_neck_cancer |
| [13] | Wang H, Kuehn A, Bouzyk E, et al. Web-based head and neck cancer tissue database American Association for Cancer Research (AACR) 2009 annual meeting poster, April 18-22, 2009, Denver, CO |
| [14] | ArgoUML Available from: http://argouml.tigris.org |
| [15] | Introduce Available from: http://cagrid.org/wiki/Introduce |
| [16] | Common Security Module Available from: http://ncicb.nci.nih. gov/NCICB/infrastructure/cacore_overview/csm |
| [17] | Langella S, Hastings S, Oster S, et al. Sharing data and analytical resources securely in a biomedical research grid environment J Am Med Inform Assoc 2008; 15(3): 363-73. |
| [18] | Head and Neck Cancer Specialized Programs of Research Excellence (HNC SPORE) Available from: http://spores.nci.nih. gov/current/hn/hn.html |
| [19] | User Provisioning Tool (UPT) Available from: https://wiki.nci.nih. gov/display/caCORE/FAQs+-+CSM+-+UPT#FAQs-CSM-UPT-CommonSecurityModuleUserProvisioningTool(UPT) |
| [20] | caCORE SDK 4.0 Developer’s Guide Available from: https://gforge.nci.nih.gov/docman/view.php/148/8650/caCORE SD K 4.0 Developers Guide_101007.pdf |
| [21] | Introduce 1.2 tutorial Available from: http://cagrid.org/wiki/ CaGrid:Tutorials:1.2:DataService:caCORE_SDK_4 |
| [22] | caTissue Suite Available from: https://cabig.nci.nih.gov/tools/catis suesuite |



