RESEARCH ARTICLE
The State of the Art of Medical Imaging Technology: from Creation to Archive and Back
Xiaohong W Gao*, 1, Yu Qian 1, Rui Hui 1, 2
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
Issue: Suppl 1
First Page: 73
Last Page: 85
Publisher Id: TOMINFOJ-5-73
DOI: 10.2174/1874431101105010073
Article History:
Received Date: 13/5/2011Revision Received Date: 17/5/2011
Acceptance Date: 17/5/2011
Electronic publication date: 27/7/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
Abstract
Medical imaging has learnt itself well into modern medicine and revolutionized medical industry in the last 30 years. Stemming from the discovery of X-ray by Nobel laureate Wilhelm Roentgen, radiology was born, leading to the creation of large quantities of digital images as opposed to film-based medium. While this rich supply of images provides immeasurable information that would otherwise not be possible to obtain, medical images pose great challenges in archiving them safe from corrupted, lost and misuse, retrievable from databases of huge sizes with varying forms of metadata, and reusable when new tools for data mining and new media for data storing become available. This paper provides a summative account on the creation of medical imaging tomography, the development of image archiving systems and the innovation from the existing acquired image data pools. The focus of this paper is on content-based image retrieval (CBIR), in particular, for 3D images, which is exemplified by our developed online e-learning system, MIRAGE, home to a repository of medical images with variety of domains and different dimensions. In terms of novelties, the facilities of CBIR for 3D images coupled with image annotation in a fully automatic fashion have been developed and implemented in the system, resonating with future versatile, flexible and sustainable medical image databases that can reap new innovations.
1. INTRODUCTION
Medical imaging has learnt itself well into modern medicine as a hallmark in the last 30 years and revolutionized health care delivery and medical industry, benefiting the world to a greatest extent, both personally and professionally. Stemmed from the discovery of X-ray by Nobel laureate Wilhelm Roentgen, radiology was born, so was the medical imaging industry. In 2010 alone, as many as 5 billion medical imaging studies were conducted worldwide [1], demonstrating the rich application of images.
Extending human vision into the very nature of disease, medical imaging is enabling a new and more powerful generation of diagnosis and intervention. For example, image-guided surgery proffers minimum invasive operations, while removing intended target completely by transforming vision into reality, not only saving lives that could not be helped otherwise but also bringing immeasurable value to countless more patients, survivors and their loved ones.
To this end, the acquired images not only serve as a barometer measuring the degrees of deterioration of a patient’s disease as well as the extent of recovering progress, but also contribute to a great deal to making a correct clinical decision. As a direct result, efficient and effective management systems are in amounting demand to ensure the data archived today will be still available tomorrow being safe and retrievable. For radiologic images, Picture Archiving and Communications Systems (PACS) are in place to manage them, whereas at the mean time many disparate databases are employed to archive domain-based data. Although plethora efforts have been made to update PACS into managing the other non-radiology images, it has been proven that the model of ‘one-size-fits-all’ is neither sustainable nor workable for today’s modern imaging facilities and image-based applications. For instance, the current state of the art radiology therapy offers highly personalized care that can be tailored to each patient’s unique characteristic factors that cannot be formulated by a unified model.
On the other hand, with this vast amount of rich collections of medical images growing from terabyte (=1012) to petabyte (=1015), innovations can be made from it, as put forward by Sir Muir Gray [2], then chief knowledge officer of the National Health Service (NHS) at the UK, the ‘application of what we know already will have a greater impact on health and disease than any drug or technology likely to be introduced in the next decade’ Image repositories or databases are a typical example collecting large amount of information waiting to be exploited. Hence effective and efficient exploitation of existing databases can provide advances to help researchers unlock the mysteries of insidious diseases and thereby can bring better treatments and cures within reach. Furthermore, melding these advances with the power of the cutting edge digital and information technology is fostering greater efficiency, quality, and value in health care.
This paper will give a summative account of the above issues and draw attention to retrieval of images of high dimensions (>2) and annotation of images in a full automatic manner, the novel development in this study. A brief account of various image modalities is given in the next Section that is then followed by the description of archiving techniques. After giving a detailed explanation of the implementation of our current repository of 2D and 3D images, MIRAGE, the potential renovations from the existing data are then addressed, preceding the conclusion.
2. THE INNOVATIVE CREATION OF MEDICAL IMAGES
2.1. Radiology Imaging
As it happened, medical imaging started with the discovery of X-radiation by Röntgen [3] in 1895 [4]. Composed of X-rays that are in a form of electromagnetic radiation, X-radiation has the ability of identifying bone structures. Consequently X-rays have been employed widely in medical imaging, in particular in the fields of dental care and chest study. To overcome the limitation that X-rays can only give 2D views, X-ray computerized tomography (CT) was subsequently invented in the 1970s by the use of computer processors to generate three-dimensional images on the inside of an object. In doing so, a series of 2D X-ray images are taken [5] from different angles in an attempt to produce detailed cross-sectional pictures of the areas inside the body. As a consequence, the resulting images can provide more information than regular X-rays, and thereforeallow doctors to look at individual slices within these 3-D images. This creation has led to the awards of Nobel Prize to the two creators, Cormack and Hounsfield, in 1979. Since then, CT has become an important tool in medical imaging to supplement X-rays and commonly referred as a CAT scan, which is applied into many fields includingchest, colon health (CT colonography), pulmonary embolism (CT angiography), abdominal aortic aneurysms (CT angiography) and spinal injuries.
Although CT has been routinely applied in the study of human brain, it is not capable for the investigation of functions, giving rise to the development of positron emission tomography (PET) and single photon emission computed tomography (SPET). PET [6] is a type of nuclear medicine imaging technique that produces a three-dimensional image or a picture of functional processes in the body, i.e., how tissues and organs are functioning. By doing so, the system detects pairs of gamma rays emitted indirectly by a positron-emitting radionuclide (tracer) on a biologically active molecule, which is introduced into the body before the scanning. Images of tracer concentration in 3-dimensional or 4-dimensional space (i.e., including time) within the body are then reconstructed by computer analysis software. Because of this advantage, PET is often used to evaluate and study neurological diseases such as Alzheimer’s and multiple sclerosis, cancer, and heart disease. In modern scanners, this reconstruction is often accomplished with the aid of a CT X-ray scan performed on the patient during the same session in order to show the bone structure of an object.
On the other hand, the arrival of magnetic resonance tomography (MR)[7] had strengthened the earlier creations in a radiology department with its first application taking place in 1977 [8]. It appears to offer better contrast and detailed internal structures by showing the difference between normal and diseased soft tissues of the body than PET or CT and contributes significantly when imaging organs like brain, muscles, heart and cancerous objects. Unlike CT scans or traditional X-rays, MR does not utilize ionizing radiation. Instead it employs a powerful magnetic field to align the magnetization of some atoms in the body, and then relies on radio frequency fields to systematically alter the alignment of this magnetization.
Fig. (1) illustrates an example of 3D datasets of brain images for the same subject obtained by different modality of imaging techniques, including CT, MR and PET. In addition, in order to retain its positive features of each image technique, hybrid tomographyis emerging by consolidating two imaging techniques, for example, PET/CT and PET/MR can display both functional and structural information within one set of constructed images.
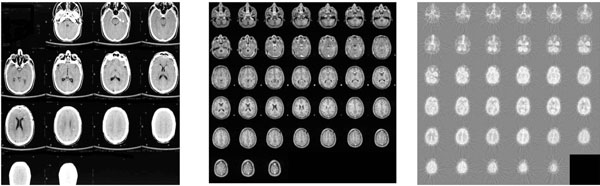 |
Fig. (1). A3D brain imagesof the same subject are acquired using CT (left), MR (middle) and PET-FDG (right). |
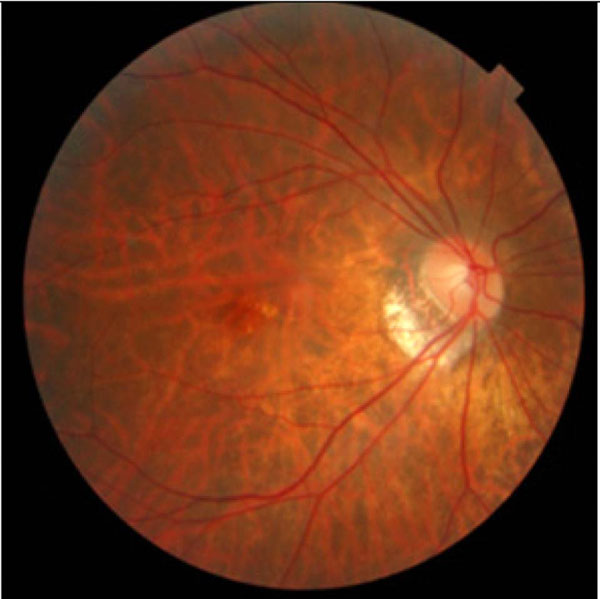 |
Fig. (2). An image showing the back of the author’s fundus. |
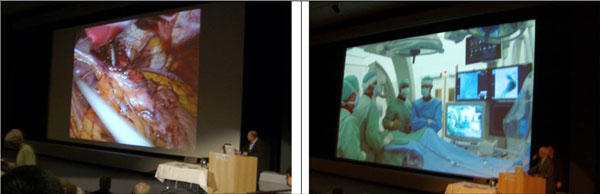 |
Fig. (3). Live operation (top) shown at EuroPACS 2006 (taking place at Trondheim, Norway) via video conferencing links (bottom) performed by Oslo University. |
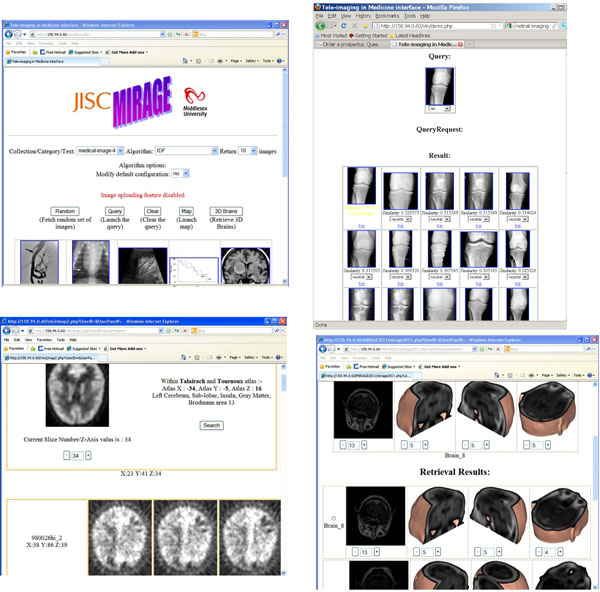 |
Fig. (4). An image retrieval system with CBIR facility at http://image.mdx.ac.uk/. Top-left, interface with menu bars; top-right, CBIR for 2D images; Bottom-left: atlas-based;and bottom-right: CBIR for 3D images, whereas the arrow pointing the four texture-based approaches that have been implemented. |
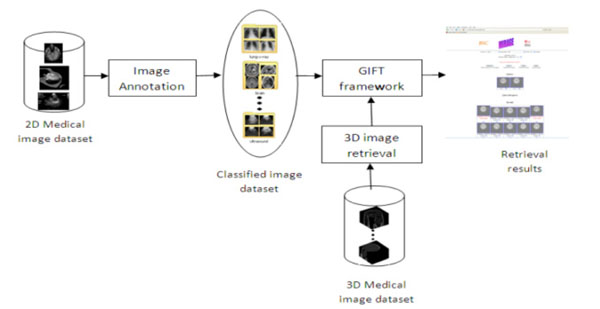 |
Fig. (5). The framework for MIRAGE. |
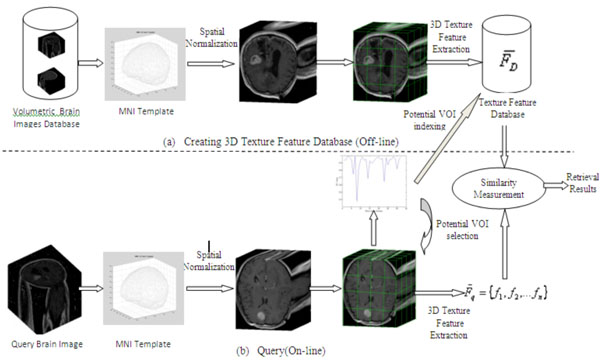 |
Fig. (6). Framework for Content-based 3D Brain Image Retrieval. |
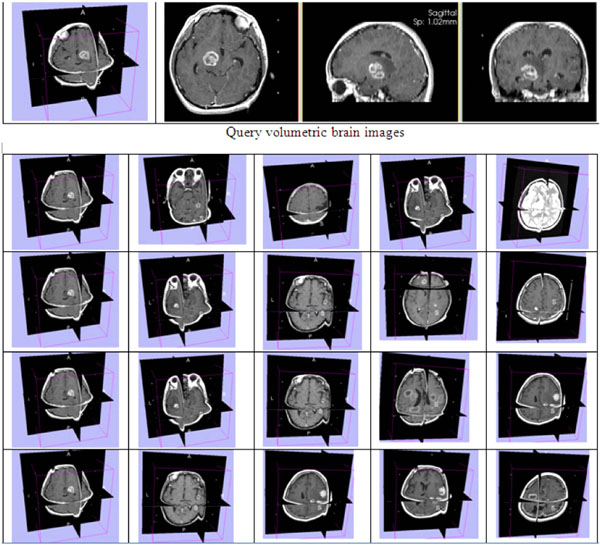 |
Fig. (7). Retrieved results in top 5 ranking from 3D GLCM (row 1), 3D WT(row 2), 3D GT (row 3), and 3D LBP (row 4). |
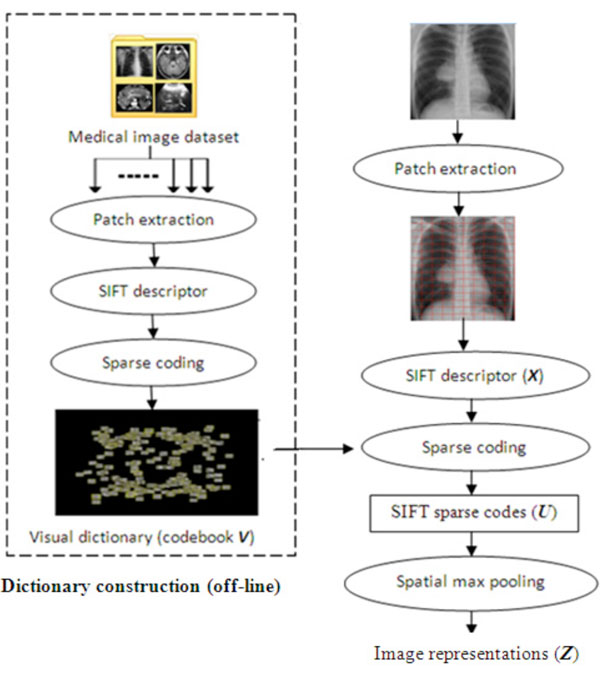 |
Fig. (8). Dictionary construction and image representations. |
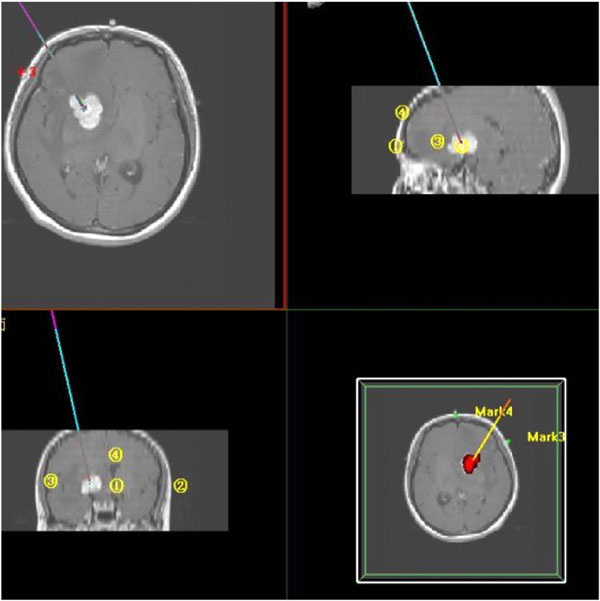 |
Fig. (9). Path planning for Image-guided neurosurgery where lines refer to the route to insert a probe which are viewed from three directions. The tumor is shown in red on the bottom-right graph. |
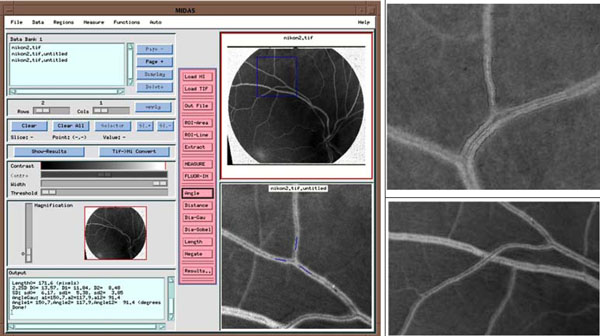 |
Fig. (10). A software system measuring vessel geometries. Top: interface with angle measurement. Middle and bottom: diameter measurement and vessel tracking to measure vessel length (black dots). |
2.2. Ultrasonic Imaging
In addition to those radiological techniques discussed in Section 2.1, there are many other imaging scanners that have been widely used as diagnostic tools in modern medicine. Diagnostic ultrasound, also known as medical sonography or ultrasonography, has been in service to image the human body for more than 50 years. It utilizes high frequency sound waves that are greater than the upper limit of human hearing to create images on the inside of a body [9] by converting the returning sound echoes into a picture. Subsequently, this technology can also produce audible sounds of blood flow, allowing medical professionals to use both sound and visual data to assess a patient’s health. The well-known application fields of ultrasonic imaging include monitoring pregnancy, studying abnormalities in the heart and blood vessels, and investigating organs in the pelvis and abdomen.
2.3. Optical Imaging
Another addition to the advanced radiological imaging technology is the optical imaging techniques that are increasingly relied on to supply traditional patient information in order to arrive at an accurate diagnostic decision. The examples include images of skin lesions in dermatology, retinal pictures in ophthalmology, endoscopic images in gastroenterology and otolaryngology, and gross and microscopic specimens in pathology. The basic principle behind these techniques is the same as that of a camera to capture the light, hence coined the name of light imaging techniques. Fig. (2) shows an example of retinal image that is taken from the pupil of an eye (the author Gao’s eye), the only window that can see vessel distributions in vivo [10].
Additionally, endoscopic imaging allows medical doctors to peer inside the human body, examining the interior of a number of organs and helping to diagnose medical conditions much earlier and more effectively in the treatment process. Although modern medical device makers have done their best to minimize the scope and size, by its very nature any form of endoscopy is going to be invasive, i.e., somehow, a camera has to find its way to go inside of a patient. Fig. (3) illustrates an example of key-hole surgery performed on an abdomen with the technique of endoscope.
3. THE STATE OF THE ART OF ARCHIVING TECHNIQUES
With such large amount of image data created every day, image management faces a huge challenge, in many ways if not possible. By nature, medical images are highly heterogeneous with a variety of domains, including brain, abdominal, cardiac, etc.. Whereas upon each domain, images can be acquired using many imaging modalities, as addressed in Section 2. Furthermore, each image dataset can be presented in various formats (DICOM, TIFF, JPEG) and in different dimensions (2D (retinal), 3D (brain), 4D (e.g, a 3D heart over a period of time), and even 5D (e.g, a functional 4D at a specific anatomic location). Thus, it is neither practical nor work able to standardize these data into a ‘one size fits all’ model. Archiving these data therefore remains a challenging but rewarding task to provide clinicians with all the necessary data in a seamless fashion at the point of care. Significantly, the established archiving system has to be with scope, scale, and the necessary flexibility, to ensure the databases established today will be still in operation tomorrow in the rapidly progressing digital era.
Although at present, large amount of images can be accommodated easily and cheaply, thanks to the advances of computer hardware technology on storage media, it is the way in which images are indexed and thereby retrieved back in real time that appears arduous, for browsing through billions of images in order to find the right one is not an option. Image retrieval can be either conducted based on text-based labelling, e.g., patient’s ID, or textual annotation that can be extracted from an image manually or semi- or full-automatically, or upon its visual or embraced content. In the later case, the retrieval is performed by querying the collections using a sample image to search for the data with similar appearance or similar features, in other words, content-based image retrieval (CBIR).At present, in clinical applications, all the medical image systems are indexed and therefore retrieved using text-based features. Specifically, for radiology images, Picture Archiving and Communications Systems, or more commonly known as PACS, has been in place to supervise the workflow by distributing, managing and interpreting images.
3.1. PACS
The first generation of the software of PACS was developed in the mid-1980s in the USA and Europe [11] when modules of PACS were programmed and were implemented independently in the departments of different domains, e.g., paediatric, coronary, or neuro-radiology. PACS enable images, such as X-ray scans, to be stored electronically and viewed on video screens, so that doctors and other health professionals can access the information and compare it with previous images at the touch of a button. The arrival of PACS has since replaced large numbers of radiological film, the medium that had been around for over 100 years and had been almost the exclusive way for capturing, storing, and displaying radiographic pictures. PACS also remove all the costs associated with hard films and releases valuable space currently used for storage. Most importantly, PACS have the potential to transform patients’ experience of the care they receive across an array of networked hospitals. By this way, if a patient has relocated from one place to another, his/her medical records together with their medical images can be easily retrieved from the system as if from the same hospital.
In principle, PACS can take forms of any of image formats, such as TIFF, or lossy compression format JPEG, as well as home grown formats built into an image scanner. In practice, however, in order to communicate images with each other to ensure an image acquired from one radiology department can be readable by the others, a standard format is required to prevent the repetitive work on converters translating formats from one to another. This requirement has led to the development of DICOM, also known as (aka) Digital Imaging and Communications in Medicine, which was initiated in 1992 in the USA, while in liaison with other Standardization Organizations in both Europe and Japan [12]. DICOM currently has widely been utilized in medical imaging field, in an attempt to facilitate standard communications of medical images. This standard has been continuously extended to meet the demands of practical applications.
3.2. CBIR
As much as we enjoy using text to search a database of any types of data due to its simplicity and straightforwardness, other forms of retrieval would be welcome for interpretation of images into words is neither nature nor practical. Although PACS has gradually matured over the last 25 years, there are still abridged parts to be bridged even for radiologic images. For example as explained in [13], general radiology PACS don't provide the display functionality to shore up nuclear medicine studies. In addition, some PACS don't fully support DICOM for nuclear medicine in favor of easy storage and retrieval of nuclear medicine studies. Nevertheless, because of the maturity of PACS, other image specialties tend to join PACS, which has led to the deployment of mini-PACS as self-contained tailor-made image management system for visible light images, for example, mini-PACS for pathology [14], by which image scanners are in service to scan specimen slides, whereas mini-PACS archive and store the scanned images. This however in turn leads to an issue of IT infrastructure since the size of this technology generated can be in an excess of 1 TB per week in a typical institution with 10 pathologists and can well overwhelm local global IT facility infrastructures. Furthermore, the challenges of integration and interoperability have to be faced when it comes to join the main flow of whole hospital PACS, as these scanning devices need to accommodate the currently used and accepted standards, including DICOM for image format and HL-7 (http://www.hl7.org/about/index.cfm), for information systems connectivity, which appears far a field. With this in mind, an efficient and effective archiving system requires a novel architecture to ensure queries can span not only to data sources of distributed collections, but also without prior standardization of the original data, entailing each local-repository with scope, scale and flexibility. Therefore, mini, niche, and third-party databases that fit for purpose should be encouraged instead of opting for a unified PACS that fits for all.
In a PACS, the search for any image is normally performed by usingtext, such as patients’ ID numbers, the types of disease, etc., which is a very efficient way in search of correct records of a patient, especially useful when it comes to interface with Hospital Information Systems (HIS) or Radiology Information Systems (RIS) with known IDs. But, the establishment of image archive systems also serves the intension of research and education, leading to new discoveries and creations. To this end, searching image databases based on contents, i.e., the information that an image contains can certainly help in an effective way in finding relevant data, especially within a repository housing large amount of image volumes where browsing through the data is impossible. Therefore a plethora amount of research has been carried out to study content-based image retrieval for the past 25 years with many promising results [15-17], in particular, since the forum of image CLEF, the cross language image retrieval [18] was established in 2003.
3.3. Content-Based Image Retrieval (CBIR) for 2D Images
Since the emerging of internet in 1990, coupled with the advances of computer hardware, vast amount of textual and imagery data are available online, prompting the creation of text-based search engine, such as Google and Yahoo, in an attempt to filter the relevant data. For image data, however, thanks to their embedded information being in the pictures, CBIR has been researched both horizontally and vertically. As the trend continues, the progress in the last century (1994-2000) has been very well documented at [19], whereas the state of the art in the last decade (2001-2008) was reviewed by [20] with future directions. In general, a CBIR system extracts features of images in terms of their global visual information, such as colour, texture, and shape, and then represents these features using mathematical vectors that in turn are employed to index each image. When a query image is submitted, the system only needs to extract these features from the query and perform the comparison with the feature database that has been stored in advance. In this way, the retrieval process of an image can be as fast as that in a text-based system since the similarity calculation is based on numerical data.
In terms of CBIR in the medical domain, the research started in 2002 with many established systems, mainly with an intention of research and illustration [21, 22]. The more recent development on 2D images has been reviewed at [23]. CBIR might not be applied in any medical domain at present; but it is the nature of utilizing images to search images which shows its potentials.
3.4. CBIR for 3D Images
With the advances of modern imaging techniques, more and more medical images are in three or higher dimensions, allowing a coherent and collective view. However most current databases still archive and index them in 2D form based on the fact that many of these images are constituted of 2D slices, especially in CBIR systems. As a result, a number of limitations have arisen with the most significant one being that the information extracted from a single 2D slice cannot be representative as slices are getting thinner, or in other words, resolutions are getting higher.
In terms of non-medical images, retrieving 3D images mainly focuses on shapes, to meet the demand of internet retrieval. For this reason, the feature of invariance of transformations (including rotation, scaling and translation) is required to ensure the same object can be retrieved when the view angle is changed [24, 25], i.e., invariant of view angles. In contrast, retrieval of medical images of 3D pays more attention to the inside of an organ rather than to the overall shape, as in the case of brain images. Therefore for medical images, the approaches employed are quite different. Table 1 lists most of the current 3D CBIR systems for medical images, which are further detailed below.
3D CBIR Systems of Medical Images
| Name/Feature | Imaging Modality | Domain | Reference |
|---|---|---|---|
| QBISM / intensity-based | MRI/PET | Brain | Arya [26] |
| Pre-defined-semantic-based | CT | Brain | Liu [27] |
| MIMS / ontology-based | All | All | Chbeir [28] |
| Knowledge-based | All | All | Chu [29] |
| ILive - modality-based | All | All organs | Mojsilovic [30] |
| 2D Texture-based | MR | Heart | Glatard [31] |
| FICBDS / Physiological information -based | Functional PET | Brain | Cai [32] |
| 3D PET / lesion-based | PET | Brain | Batty [33, 34] |
| MIRAGE / 3D texture-based | MR | Brain | Gao [24], Qian [25] |
VOI Detection Rate
| VOI Location | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Total |
| Number of images | 24 | 46 | 18 | 38 | 24 | 12 | 14 | 8 | 184 |
| Correctly detected images | 24 | 42 | 16 | 34 | 24 | 8 | 12 | 8 | 168 |
| Correct Detection Rate (%) | 91.3 |
Retrieval Time and Precision of 3D CBIR
| Mean Retrieval Time | Mean Average Precision | |||
|---|---|---|---|---|
| Methods | Without VOI Selection | With VOI Selection | Without VOI Selection | With VOI Selection |
| 3D GLCM | 43.37s | 10.96s | 0.677 | 0.690 |
| 3D WT | 4.46s | 1.22s | 0.731 | 0.749 |
| 3D GT | 38.79m | 10.77m | 0.714 | 0.691 |
| 3D LBP | 0.74s | 0.21s | 0.774 | 0.786 |
Confusion Matrix for the Six Medical Image Categories, where B, L, M, A, U, G Represent Categories Of Brain, Lung, Microscopy, Abdomen, Ultrasound, and Graph
| Classification Results | AR (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| B | L | M | A | U | G | |||
| Ground Truth | Brain | 48 | 0 | 2 | 0 | 0 | 0 | 96 |
| Lung (X-Ray) | 0 | 50 | 0 | 0 | 0 | 0 | 100 | |
| Microscopy | 0 | 0 | 49 | 0 | 1 | 0 | 98 | |
| Abdomen | 0 | 0 | 0 | 50 | 0 | 0 | 100 | |
| Ultrasound | 0 | 0 | 0 | 0 | 50 | 0 | 100 | |
| Graph | 0 | 0 | 0 | 0 | 0 | 50 | 100 | |
| ER(%) | 0 | 0 | 3.92 | 0 | 1.96 | 0 | ||
In the system of QBISM, 3D functional brain images are queried and visualized [26], by which intensity-based volume data are stored for spatial references, whereas brain atlas Talairach [35] is employed to concert a region-based retrieval. The key to the system is the application of volumetric data type (i.e., Region or Volume as expressed as <x, y, z, value>) to represent image data.
In other cases, the retrieval task of 3D images can work well based on the feature extraction [27] from 2D slices, depending on the application fields of the created database.
On the other hand, semantics based retrieval may cover images of all dimensions as evidenced by [30]. The strength of this work therefore lies in the approaches employed for categorisation of images that bear semantically well-defined sets. In other words, the images should be represented by well-defined combinations of features with varying levels (low, high, global, regional, etc.). This task itself however in most cases poses greater challenges than semantic representation itself. Nevertheless, semantics based retrieval of medical images remains one of the current trends. Likewise, ontology-based [28] and knowledge-based approaches [29] can shorten the semantic gap to a certain extent between low level features and high level semantics, which in turn requires skilful expertise, i.e., knowledge, to interpret images and convert contents into textual descriptions.
For subject-based images that bear centralised characterises, local features can play an important role in indexing and retrieving images. For example, the system of FICNDS [32] employs physiological kinetic features for retrieving images. Similarly, Batty and Gao [33, 34] have employed binding potential (BP) values to index functional PET images. Although effective, this method is very discipline-defined and heavily relies on the additional supply of extra information. In FICNDS, to define a tracer kinetic model, plasma time activity (PTA) curves should be obtained from a series of blood samples, which are not easily available for most of the images in a database. Although PTA can still be modeled by the application of control regions as applied by Gao et al, a sequence of images acquired over a period of time, say 90 minutes, are still in need, which again is not readily available in most of image repositories. Additionally, in essence, the establishment of kinetic models stems from the data of 2D slices, which may lose information in between slices. For a system warehousing images of variety of domains, more general approach appears to be in demand in order to be sustainable.
Texture-based approach for retrieving of 3D+ cardiac images has been applied by Glatard [31] with the employment of 2D Gabor filter. While working on 4D (3D + time) heart images, their adoption of Gabor is again a strict 2D form based on regions coupled with an extra parameter dedicated to myocardium feature.
Although a texture-based approach can be applied in a 2D form due to the nature that a 3D dataset is composed of a stack of 2D slices, using a slice-by-slice 2D approach can miss the information that interlaces within the volumetric data. Thus, in terms of a 3D form of texture, this spatial structural information should be extracted from a cube instead of a square. Towards this end, while working on images of 3D brain, Gao [24] and Qian [25] have extended the approach of Local Binary Pattern (LBP) [36] into 3D form to extracting texture information that is subsequently utilized for indexing them in their developed system MIRAGE (http://image.mdx.ac.uk/vin/demo.php), which is described in details in the next Section. The other three popular texture based approaches, including Grey Level Co-occurrence Matrices (GLCM), Wavelet Transforms (WT) and Gabor Transforms (GT), are also implemented in the system, again with appropriate extensions to 3D forms.
4. MIRAGE - AN ONLINE LEARNING SYSTEM ON MEDICAL INFORMATICS
Fig. (4) depicts a few interfaces of a CBIR system, MIRAGE. With the server located at Middlesex University in the UK, the system at present accommodates over 100,000 2D and 3D images and facilitates domain-based(top-right), atlas-based(bottom-left), and content-based retrieval for both 2D and 3D images (bottom-right).
4.1. The System
Fig. (5) illustrates the flowchart of the architecture of MIRAGE. To address the problems that current text-based image retrieval systems suffer, MIRAGE integrated the methods of both CBIR for 2D and 3D collections and automatic image annotation to label the images with its keywords, leading to a high level of semantic search. It is therefore consists of three modules as shown in Fig. (5), with components of image annotation, 2D image retrieval and 3D image retrieval respectively.
Built on the open source GNU Image Finding Tool (GIFT, http://www.gnu.org/software/gift/), the online database is based on the Query-by-Example (QBE) paradigm coupled with a facility of user-relevance feedback whereby retrieved images most closely resemble a query image in appearance (i.e., the content that an image is carrying).
For 2D images, two algorithms have been implemented for indexing image collections, which are IDF (Inverse Document Frequency) and Separate Normalization. The IDF is a classical method and is based on counting the number of documents in the collection being searched, which contain (or are indexed by) the terms in question [37], and has been applied in text retrieval systems, giving rise to the efficiency when employed in an image system.
Additionally, feature normalization is required to compensate the scale disparity between the feature components that are defined in different domains. On the client side, a web page based interface is given. Whilst the client-server communication is achieved using the XML-based Multimedia Retrieval Mark up Language (MRML). All client-server communication, including queries from the client or results returned by the server, is realized through message passing. Consequently, the client can be implemented in any programming language. The current MIRAGE client is implemented using PHP (Personal Home Programming) language to generate dynamic web pages for the client web browser.
4.2. Archiving 3D Brain Images in MIRAGE
To develop a CBIR system, two phases are in need including deposit and retrieval respectively. At the phase of deposit of the data, the collected data firstly undergo a pre-processing stage to normalize them to ensure that all the images share the same resolution before the indexing stage.
For 3D brain images, four texture based methods are implemented as pointed in Fig. (4, at bottom-right graph), including 3D Local Binary Pattern (LBP), 3D Grey Level Co-occurrence Matrices (GLCM), 3D Wavelet Transforms (WT) and 3D Gabor Transforms (GT) that are detailed in [24, 25]. Fig. (6) displays the framework of CBIR for 3D images.
As illustrated in Fig. (6), the collection of 3D brain images firstly underwent a pre-processing stage to normalize them into the same resolution before the indexing stage. After spatial normalization of volumetric brain data into a standard template, the data are then divided into 64 non-overlapping equally sized blocks, from which, 3D texture features are extracted to create a feature database. On the query side, a pre-processing stage is introduced to detect the potential VOI of lesions after spatial normalization from a query image. Subsequently, the extraction of 3D texture features from a query only takes place from these potential sub-blocks that, in the retrieval stage, are in turn compared with the corresponding features in the feature database to obtain retrieval results.
Specifically, the formulae of 3D forms for the four texture-based approaches are an extension of their 2D versions. For example, in terms of 3D LBP, a 3D dynamic texture is recognized by concatenating three histograms obtained from the 2D LBP on three orthogonal planes, i.e., XY, XZ, and YZ planes, while applied to our normalized brain images and is expressed as
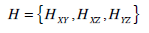 |
(1) |
where
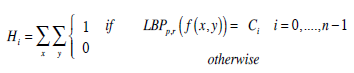 |
(2) |
And LBPp,r (f (x, y)) is LBP operator in 2D form, and is derived from a general definition of texture in a local circular (p, r) neighborhood. In other words, for each pixel in an image f (x,y), a binary code Ci is produced by thresholding its value with the value of a centre pixel.
On the other hand, in 3D form, GLCM are defined as two dimensional matrices of the joint probability of occurrence of a pair of grey values separated by a displacement d = (dx, dy, dz), which can be calculated using Eq. (3).
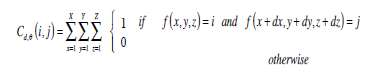 |
(3) |
where d,θ represent the offsets of distance and angle in order to achieve rotation invariant and subsequently determine the values of dx, dy, and dz. A normalized co-occurrence matrix Pd,ϑ(i, j) is usually obtained by
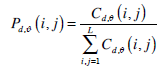 |
(4) |
where L is the total number of grey level of a 3D image.
In addition, a 3D WT provides a spatial and frequency representation of a volumetric image and is formulated as Eq. (5) for a 3D image of f (x,y,z).
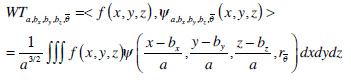 |
(5) |
where 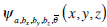 is the daughter wavelet generated by translation on xy and z-axes by bx, by and bz respectively, dilation by a, and rotation by the angle
is the daughter wavelet generated by translation on xy and z-axes by bx, by and bz respectively, dilation by a, and rotation by the angle 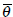 of the mother wavelet ψ (x,y,z).
of the mother wavelet ψ (x,y,z). 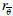 refers a rotate factors in 3 dimensional space.
refers a rotate factors in 3 dimensional space.
With respect to Gabor Transforms, Eq. (6) has been applied in MIRGAE as a 3D Gabot filter.
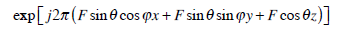 |
(6) |
where 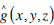 is a 3D Gaussian function, together with radial centre frequency F and orientation parameters(θ and φ), determining a Gabor filter in three dimensions. Given a 3D volumetric textured image f (x,y,z), its 3D Gabor transform GTi is then defined by
is a 3D Gaussian function, together with radial centre frequency F and orientation parameters(θ and φ), determining a Gabor filter in three dimensions. Given a 3D volumetric textured image f (x,y,z), its 3D Gabor transform GTi is then defined by
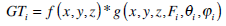 |
(7) |
Fig. (7) demonstrates an example of retrievedusing different texture approaches.
Similar to the situation in 2D forms, the judgement of each approach is subject to the applications of the retrieval task as to which of the measures of size, location or shape plays more important role than the others.
4.3. Image Annotation
Another feature that the MIRAGE has is its ability of annotating imagesin a fully automaticfashion, in ahope to achieve ahigh level search ofsemantics while organizing and categorizing images of interests. Automatic image annotation is the process by which a computer system automatically assigns metadata that arecaptioning or keywords to describe a digital image. At present, the Bag-of-visual-Words (BoW) [38] paradigm becomes very popular and has been successfully applied in image categorization. By transforming images into a set of symbolic ‘visual vocabulary’, images are represented using the statistics of the appearance of each word as feature vectors, upon which the learning of an image classification rule could be achieved by a classifier. This idea has been adopted in the MIRAGE system coupled with SIFT sparse coding approach [39], which is conducted in the following four steps as indicated in Fig. (8).
- Step 1 -- the visual features are extracted from local patches of each image in the training dataset, leading to the construction of a visual dictionary of codebook;
- Step 2 -- to quantize the visual features of the image dataset into discrete symbolic‘visual words’;
- Step 3 -- an image is represented as a unique distribution (e.g. a histogram) over the generated dictionary of words; and
- Step 4 -- image representations of the training dataset obtained in Step 3 are applied to train the classifiers using supervised machine learning methods, thereby the trained classifier automatically allocates new images into corresponding categories and hence labels them.
Different from traditional BoW paradigm, sparse coding is employed in the MIRAGE instead of vector quantization (VQ) to extract the SIFT descriptors of local image patches. Furthermore, instead of using histograms, multiple scales of max pooling are utilized as an image representation by the application of simple linear support vector machines (SVMs). This is in the consideration that when compared with the SVMs using nonlinear kernels of histogram intersection, linear SVMs can dramatically reduce the training complexity while maintaining a good performance.
4.4. Evaluation Resultsof theMIRAGE
4.4.1. Results on CBIR
In terms of retrieved results with reference to lesion locations, LBP approach appears to perform the best amongst the four texture-based methods. Table 2 gives the retrieval results on the evaluation of LBP, where by the first row is the labeling number of the location of a VOI assigned by the authors for the convenience of calculations, e.g., ‘1’ refers to the abnormal part in the front top left-most part of the brain. The second row is the total number of images containing VOIs in such positions in the database, whilst the number of correctly detected images by the LBP is given on the third row. Therefore the overall performance of the LBP with respect to VOI locations is measured as a ratio of detected positive VOIs to the total positive VOIs, yielding 91.3% (=168/184).
In terms of precision, the retrieved accuracy is 78% based on ten query images with the ground truth being diagnostic information on the locations and sizes of tumors, demonstrating very promising results.
It is understandable that retrieving images in 3D form might not be performed in real time due to its lengthy processing time occurred on whole volume of data rather than a single 2D slice, one of the setbacks in the development of CBIR systems for higher dimensions. Table 3 lists the average querying time, accounting for the total time that has spent on both feature extraction and retrieval. The second column is the averaged querying time without a pre-processing stage of VOI selection, whilst the third column is with VOI selection. All methods run in Matlab R2009a with a CPU of an Intel P8600 1.58GHz and 3.45GByte RAM.
As can be seen in Table 3, the retrieval time with VOI selections offers 4 times faster than the one without. In particular, the retrieval time for 3D GT takes a much longer time than the other methods spending 38 minutes, due to the employment of 144 times of 3D convolutions for each block, whereas the query times for the other methods are in the space of few seconds. The table is also in favor of 3D LBP approach with sub-second retrieval times and the highest precision rate of 78%.
4.4.2. Results on Image Annotation
To assess the effectiveness of image classification, a confusion matrix is firstly created as explained at [40]. In addition, in order to train a codebook for image annotation, a training dataset is firstly selected containing 1000 images that are randomly chosen from our medical image repository. Then 200,000 random patches are collected from these images. Subsequently, from each patch, SIFT descriptors are extracted, yielding a feature database that has the size of 200,000*128 elements, which are finally applied to train the codebook with the size of 1024*128 in terms of feature vectors.
With respect to ground truth for image annotation, six domain names are defined at the highest level, which are brain, lung (x-ray), microscopy, abdomen, ultrasound and graph. Each category is allocated 100 images as ground truth with each half (50) as being training and testing sets respectively. The classification results for the six categories are visualized in a confusion matrix given in Table 4.
The values in the last column of Table 4 are the Accuracy Rate (AR) values for each class, whereas the values in the last row are the Error Rate(ER) for each class. The Average Accuracy Rate (AAR) for all classes is 99% (297/300), and Average Error Rate (AER) is 1% (3/300), demonstrating the approach of annotation being of efficiency though not conclusive. In addition, an online questionnaire has been created to help to further improve the system. So far, the feedbacks have not shown any untoward result with over 70% response in ‘strong agreement’ with ‘retrieved results’, ‘ease to use’ and ‘useful’ in e-learning processing.
5. DISCUSSION - IMAGE INFORMATICS
With such wonderful creation of imaging techniques and comprehensive archiving systems fostering generated huge amount of image data, it is anticipated that more valuable information can be exploited, leading to more creations.
A database serves two ways, which is from creation to archive and then from archive to creation. Therefore it is in the expectation that the collection of these huge volumes of medical images can help researchers to unlock the mysteries of insidious diseases and thus can bring better treatments and cures within reach (e.g., personalized radiation therapy). These innovations can be explored in one of the following trends that are shown as following examples to elicit more creations.
5.1. PACS-Based Informatics
Based on PACS, computer aided diagnosis are in demand as described in [41]. Towards this, medical imaging informatics infrastructure (MIII) has emerged focusing on existing PACS resources and their images as well as related data for organized horizontal and longitudinal clinical, research, and education applications which would otherwise not be able to perform due to insufficient information [42].
PACS-based imaging informatics is the next research and development frontier in utilizing PACS-related technologies and data for various image-assisted applications. These include image-assisted radiology and surgery, minimally invasive surgery, robotic and remote surgery, maxillofacial imaging, cardiovascular imaging, image-assisted detection (CAD) and diagnosis (CADx), image matching, and imaged-based learning and teaching.
5.2. Image-Guided Surgery
Image-guided surgery is another burgeoning field by using minimum invasive ways in assisting surgeons manipulating, monitoring, and/or guiding operations with the advantages of being higher precision of targeting, persistence of longer duration, and the ability of pre-operative planning drawing from patients’ images. As of 2001, there are around 270,000 cases of image-guided robotic operations conducted per year Worldwide [43]. And the trend follows.
During an image-assisted or guided surgery, the images can be acquired in real time to provide an augmented view to the surgeons as exemplified in Fig. (3), whereby a tiny camera has to be inserted into the intended position inside the body. Another image-assisted surgery or more commonly known as image-guided surgery is less invasive, especially when it is employed to perform on a brain where every tissue is too delicate to be traumatized in order ascertain a person’s normal life. In this case, images are acquired pre-operative by using MR or CT, whereby the position to open a hole on the skull, the route to insert a probe and the extent of a lesion to be incised can be planned in advance, leading to a success clinical outcome. Fig. (9) exhibits an interface of path planning where each line refers to the probe route viewed at a different angel to be inserted in for resection of the tumor (red in the bottom-right graph).
The biggest advantage of this type of minimum invasive operation is the maximum safety and minimum trauma to a patient. In most cases, as soon as an operation is completed taking about 20 minutes, patients can be back to normal, resulting in less suffering in terms of time, money, and psychological issues with none or less scars incurred.
5.3. Image-Assisted Diagnosis
With respect to optical imaging techniques, one application field is to help clinicians in making a correct clinical decision. At [10], an accurate measurement system, MIDAS, has been developed in determining vascular network properties such as power losses, shear stresses, etc., leading to the diagnosis of hypertension diseases as demonstrated in Fig. (10), whereas the left graph is the interface that assists clinicians in measuring vessel geometric in terms of angle, diameter (right-top) and vessel length (bottom-right) in-between bifurcations.
5.4. Image-Based Learning and Teaching
A picture is worth a thousand of words, which is particularly true when it comes to learning and education. In particular, in the medical domain, the study of the anatomy of internal organs of the human body relies heavily on graphs and images. Therefore, with the advances of modern imaging techniques, image-based learning and teaching have been flourished. In the case of MIRAGE [24, 25], as pointed out in Fig. (4), it has been developed to dedicate to teaching and learning, specifically for MSc students who are on the programme of Biomedical Modeling and Informatics running at Middlesex University at the UK. The system has guided students significantly, especially when they are in need to know more about anatomy of one particular organ.
6. CONCLUSION
While the wonderful creation of medical imaging techniques has benefited both patients and health professionals tremendously, it has great potentials to contribute more in finding better treatments in the future. To this end, the archive of those generated medical images of huge quantities in a sustainable way becomes paramount, especially which can be further exploited when a new generation of data analysis tools or new information become available. At present, it had been proved that the model of ‘one size fits all’ is neither sustainable nor workable, leading to the exploitation of the development of each individual database that fits for purpose and equips with facilities that can be both functioning independently and searchable when integrated with the other image databases. With this in mind, the infrastructure of the future archive systems should be lightweight, incremental and in a measured way. To this end, MIRAGE repository has been developed facilitating domain-based, atlas-based and content-based retrieval with an aim of e-learning and e-teaching, which has made huge impact to both students and lecturers. Significantly, 3D images are included together with 3D visualization over the platform of internet while delivering the learning materials. Another advantage of MIRAGE is that it employs open source software that is accessible and will contribute to open source collection of software, leading to a wider access of medical image repositories on the search of better education for students in the short-term and better solutions and better treatments for patients in the long-term.
ACKNOWLEDGEMENT
The authors would like to acknowledge JISC (www.jisc.ac.uk) at the UK for their continuously financial support for the two projects of MIRAGE and MIRAGE 2011, and thank European Commission (EC) for their funded project WIDTH under FP7 People Scheme.
CONFLICT OF INTEREST
None declared.
REFERENCES
| [1] | Roobottom CA, Mitchell G, Morgan-Hughes G. Radiation-reduction strategies in cardiac computed tomographic angiography Clin Radiol 2010; 65(11 ): 859-67. |
| [2] | Gray M. MEDINFO http://www.chi.unsw.edu.au/CHIweb.nsf/page/Conference%20Papers 2007. |
| [3] | Spiegel PK. The first clinical X-ray made in America—100 years Am J Roentgenol 1995; 164(1 ): 241-3. |
| [4] | Novelline R. Squire's Fundamentals of Radiology 5th. 1997. |
| [5] | Herman GT. Fundamentals of Computerized Tomography: Image Reconstruction from Projection. 2nd. Springer 2009. |
| [6] | Bendriem B, Townsend DW, Eds. The theory and practice of 3D PET. London: Kluwer Academic Publishers 1998. |
| [7] | Lauterbur PC. Image formation by induced local interactions: examples of employing nuclear magnetic resonance Nature 1973; 242: 190-. |
| [8] | Damadian R, Goldsmith M, Minkoff L. NMR in cancer: XVI. Fonar image of the live human body Physiol Chem Phys 1977; 9: 97-100. |
| [9] | Robertson VJ, Baker KG. A Review of therapeutic ultrasound: effectiveness studies Physical Therapy 2001; 81(7 ): 1339-50. |
| [10] | Gao XW, Bharath A, Stanton A, Hughes A, Chapman N, Thom S. Quantification and characterization of arteries in retinal images Comput Method Program Biomed 2000; 63(2 ): 133-46. |
| [11] | Huang HK, Taira RK. Infrastructure design of a picture archiving and communication system Am J Roentgenol 1992; 158: 743-9. |
| [12] | NEMA. Digital Imaging and Communications in Medicine (DICOM) Virginia 2004. |
| [13] | Lisa F. Nuclear medicine & image management: A world apart. Health Imaging & IT Available at http://www.healthimaging.com/index.php 2006 June; |
| [14] | Koller JS, Lindemann GW. Visible light: a few blips in digital Imaging for pathology, Imaging Technology News, January/ February Available at http://itn.reillycomm.com/node/28068/ 2006. |
| [15] | Gao XW, Müller H, Loomes M, Comley R, Luo S, Eds. Lecture Notes in Computer Science, 4987. Springer 2008. |
| [16] | Müller H, Gao XW, Luo S, Eds. Special Issue of Computer Methods and Programs in Biomedicine. Elsevier 2008; 92.(3) |
| [17] | Müller H, Gao XW, Lin C, Eds. Special Issue of Computerised Medical Imaging and Graphics 2006; 30(6-7 ) |
| [18] | mageCLEF, http://www.imageclef.org/ [Retrieved May 2011]; |
| [19] | Smeulders AW, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years IEEE Trans Pattern Anal Mach Intel 2000; 22(12 ): 1349-80. |
| [20] | Datta R, Joshi D, Li J, Wang J. ACM Computing Surveys 2008; 40(2 ) Article 5 |
| [21] | Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications - clinical benefits and future directions Intel J Med Inform 2004; 73(1 ): 1-23. |
| [22] | Lehmann T, Güld M, Thies C, et al. Content-based image retrieval in medical applications Methods Inf Med 2004; 43: 354-61. |
| [23] | Zhou XS, Zillner S, Moeller M, Zhan Y, Krishnan A, Gupta A. Semantics and CBIR: a medical imaging perspective In: Proceedings of the 2008 International Conference on Content-based Image and Video Retrieval; 2008. |
| [24] | Gao XW, Qian Y. Texture-based 3D image retrieval for medical applications In: Proceedings of IADIS e-Health; Freiburg, Germany. 2010. |
| [25] | Qian Y, Gao X, et al. Content-based retrieval of 3D medical images 2011 February; In: The Third International Conference on eHealth, Telemedicine, and Social Medicine (eTELEMED 2011); Guadeloupe, France. 2011. |
| [26] | Arya M, Cody W, Faloutsos C, Richardson J, Toya J. QBISM: Extending a DBMS to Support 3D Medical Images Proceed Tenth Intern Conference Data Engin 1994. |
| [27] | Liu Y, Dellaert F. A classification based similarity metric for 3D image retrieval Proceed Comput Vision Pattern Recognition 1998. |
| [28] | Chbeir R, Amghar Y, Flory A. MIMS: a Prototype for medical image retrieval Recherche d Informations Assistee par Ordinateur 2000. |
| [29] | Chu W, Hsu C, Cardenas A, Taira R. Knowledge-based image retrieval with spatial and temporal constructs IEEE Transact Knowledge Data Engin 1998; 10(6 ): 872-8. |
| [30] | Mojsilovic A, Gomes J. Semantic based categorization, browsing and retrieval in medical image databases Proceed Image Processing 2002. |
| [31] | Glatard T, Montagnat J, Mgnn IE. Texture based medical image indexing and retrieval: application to cardiac imaging Proceedings of the 6th ACM SIGMM International Workshop on Multimedia information retrieval. |
| [32] | Cai W, Feng D. Contene-based retrieval of dynamic PET functional images IEEE Transact Informat Technol Biomed 2000; 4(2 ): 152-8. |
| [33] | Batty S, Fryer T, Clark J, Turkheimer F, Gao XW. Extraction of physiological information from 3D PET brain images Proceed Visualization Imaging Image Processing 2002. |
| [34] | Batty S, Clark J, Fryer T, Turkheimer F, Gao XW. Towards archiving 3D PET brain images based on their physiological and visual content In: Proceedings of International Conference on Diagnostic Imaging and Analysis; Shanghai, China. 2002. |
| [35] | Talairach J, Tournoux P. Co-planar Stereotactic Atlas of the Human Brain. Stuttgart: Thieme 1988. |
| [36] | Unay D, Ekin A, Jasinschi RS. Medical image search and retrieval using local patterns and kit feature points In: Proceedings ofthe International Conference on Image Processing; California, USA. 2008. |
| [37] | Robertson S. Understanding inverse document frequency: on theoretical arguments for IDF J Document 2004; 60(5 ): 503-20. |
| [38] | Viitaniemi V, Laaksonen J. Spatial extensions to bag of visual words, VIVR’09. Greece: Santorini 2009. |
| [39] | Yang J, Yu K, Gong Y, Huang T. Linear spatial pyramid matching using sparse coding for image classification In: Proceedings of Conference on Computer Vision and Pattern Recognition; Miami, USA. 2009. |
| [40] | Kohavi R, Provost F. Editorial for the special issue on application of machine learning and the knowledge of discovery process Machine Learning 1998; 30: 271. |
| [41] | Huang HK. PACS-based imaging informatics: past and future PACS-based imaging informatics: past and future, International Congress Series 1256 2003; xvii-xx 2003. |
| [42] | Huang HK, Wong STC, Pietka E. Medical image informatics infrastructure design and applications Med Inform 1997; 22(4 ): 279-89. |
| [43] | Joskowicz L. Advances in image-guided targeting for keyhole neurosurgery In: Touch Briefings Reports, Future Directions in surgery. London, UK 2006; II. |



